Sample Files
Model Configs
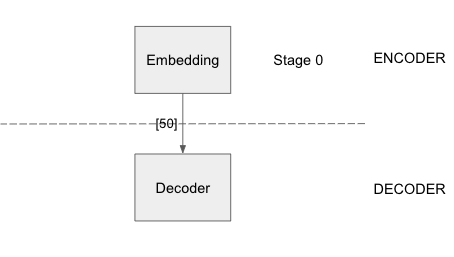
DistMult
model:
learning_task: LINK_PREDICTION
encoder:
layers:
- - type: EMBEDDING
output_dim: 50
bias: true
init:
type: GLOROT_NORMAL
decoder:
type: DISTMULT
loss:
type: SOFTMAX_CE
options:
reduction: SUM
dense_optimizer:
type: ADAM
options:
learning_rate: 0.01
sparse_optimizer:
type: ADAGRAD
options:
learning_rate: 0.1
|
|
The above configuration has a simple embedding layer whose output is fed to the decoder layer, which uses a SoftmaxCrossEntropy loss function to optimize the loss value. An Adagrad sparse optimizer is used for the node embeddings and Adam Optimizer for all other model parameters.
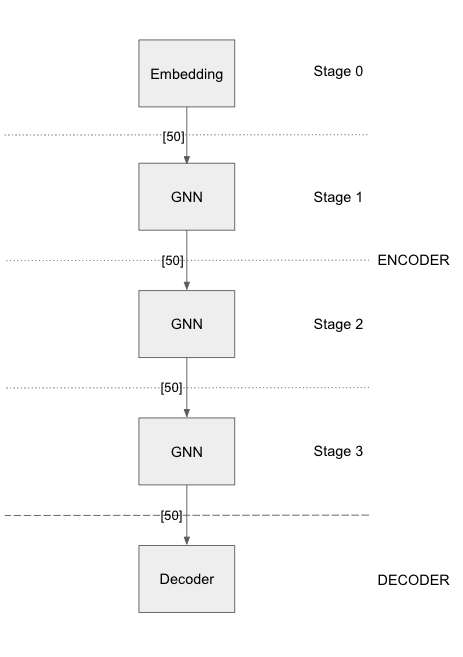
Graph Sage (3-layer)
Graph Sage (3 layer) has an initial stage consisting of an embedding layer. It is connected to 3 stages of GraphSage GNN layers. The number of training/evaluation neighbor sampling layers is equal to the GNN stages defined in the model.
GAT (3-layer)
model:
learning_task: LINK_PREDICTION
encoder:
train_neighbor_sampling:
- type: ALL
- type: ALL
- type: ALL
layers:
- - type: EMBEDDING
output_dim: 50
bias: true
init:
type: GLOROT_NORMAL
- - type: GNN
options:
type: GAT
input_dim: 50
output_dim: 50
bias: true
init:
type: GLOROT_NORMAL
- - type: GNN
options:
type: GAT
input_dim: 50
output_dim: 50
bias: true
init:
type: GLOROT_NORMAL
- - type: GNN
options:
type: GAT
input_dim: 50
output_dim: 50
bias: true
init:
type: GLOROT_NORMAL
decoder:
type: DISTMULT
loss:
type: SOFTMAX_CE
options:
reduction: SUM
dense_optimizer:
type: ADAM
options:
learning_rate: 0.01
sparse_optimizer:
type: ADAGRAD
options:
learning_rate: 0.1
GAT (3 layer) has an initial stage consisting of an embedding layer. It is connected to 3 stages of GAT GNN layers. The number of training/evaluation neighbor sampling layers is equal to the GNN stages defined in the model.
Embeddings + Features + Edges
The supported storage backends for embeddings and features are PARTITION_BUFFER, DEVICE_MEMORY and HOST_MEMORY. For edges, the supported backends are FLAT_FILE, DEVICE_MEMORY, HOST_MEMORY.
Storage Configs
GPU Memory
storage:
device_type: cuda
dataset:
dataset_dir: /home/data/datasets/fb15k_237/
edges:
type: DEVICE_MEMORY
options:
dtype: int
embeddings:
type: DEVICE_MEMORY
options:
dtype: float
In the above configuration, both edges and embeddings are stored in GPU memory.
Mixed CPU-GPU
storage:
device_type: cuda
dataset:
dataset_dir: /home/data/datasets/fb15k_237/
edges:
type: HOST_MEMORY
options:
dtype: int
embeddings:
type: HOST_MEMORY
options:
dtype: float
This configuration places the edge data in the CPU memory and maintains the embedding data in GPU memory.
Disk-Based
storage:
device_type: cuda
dataset:
dataset_dir: /home/data/datasets/fb15k_237/
edges:
type: FLAT_FILE
options:
dtype: int
embeddings:
type: DEVICE_MEMORY
options:
dtype: float
In this configuration, the edge data is stored in a flat file, on disk. FLAT_FILE storage backend is supported for edges alone, because there is no need for an index lookup. Instead, edges are traversed sequentially.
Marius supports PARTITION_BUFFER mode to store embedding data, where all data is stored on disk and only necessary chunks are fetched and kept in the buffer. The edges are traversed in an order that minimizes bukcet swaps in the buffer. It can be configured as follows
storage:
device_type: cuda
dataset:
dataset_dir: /home/data/datasets/fb15k_237_partitioned/
edges:
type: FLAT_FILE
options:
dtype: int
embeddings:
type: PARTITION_BUFFER
options:
dtype: float
num_partitions: 10
buffer_capacity: 5
The above configuration states that at most 5 node embedding buckets can be present in memory at any given time.
Training Configs
Synchronous Training
To speed up training, Graph Learning systems use pipelined architecture and try to overlap data movement with computation. This introduces bounded staleness in the system, wherein after a set of updates to the node embeddings, the existing mini-batches in the pipeline use stale node embeddings. Marius provides an explicit option to turn off asynchronous training and ensure that every mini-batch sees the latest updated node embeddings. The following can be used the set training as synchronous
training:
batch_size: 1000
negative_sampling:
num_chunks: 10
negatives_per_positive: 10
degree_fraction: 0
filtered: false
num_epochs: 10
pipeline:
sync: true
Pipelined Training
Marius uses pipelining training architecture that can interleave data access, transfer, and computation to achieve high utilization. This introduces the possibility of a few mini-batches using stale parameters during training. Below is a sample configuration where the training is async, and the staleness is set to 16 i.e. at most 16 mini-batches use stale node embeddings after any set of node embeddings are updated.
pipeline:
sync: false
gpu_sync_interval: 16
gpu_model_average: true
staleness_bound: 16
batch_host_queue_size: 4
batch_device_queue_size: 4
gradients_device_queue_size: 4
gradients_host_queue_size: 4
batch_loader_threads: 4
batch_transfer_threads: 2
compute_threads: 1
gradient_transfer_threads: 2
gradient_update_threads: 4
Marius follows a 5-staged pipeline architecture, 4 of which are responsible for data movement and the other is for model computation and in-GPU parameter updates. The pipeline field has options for setting thread counts for each of these stages.
Evaluation Configs
Link Prediction Filtered
An Evaluation configuration for Link Prediction with a batchsize of 1000. When filtered is set to true, false negative sampled edges will be filtered out.
evaluation:
batch_size: 1000
negative_sampling:
num_chunks: 1
negatives_per_positive: 1000
degree_fraction: 0.0
filtered: true
pipeline:
sync: true
epochs_per_eval: 1
Link Prediction Unfiltered
Unfiltered Evaluation configuration for Link Prediction with a batchsize of 1000. False negative sampled edges will not be filtered out.
evaluation:
batch_size: 1000
negative_sampling:
num_chunks: 10
negatives_per_positive: 100
filtered: false
pipeline:
sync: true
epochs_per_eval: 1
Node Classification
Sample Evaluation configuration for a Node Classification tasks.
evaluation:
batch_size: 1000
pipeline:
sync: true
epochs_per_eval: 1